AI for Natural Product Drug Discovery
Abstract
Artificial intelligence (AI), particularly machine learning and deep learning models, is increasingly applied in natural product-based drug discovery. This review highlights key areas where AI models—such as generative transformers and predictive algorithms—contribute to the field: genome mining for biosynthetic gene clusters (BGCs), metabolome analysis, structure prediction of natural products, and the identification of biological targets and pharmacological activities. Special attention is given to the critical bottleneck of training data quality. The lack of large-scale, well-annotated, and standardized datasets significantly limits model accuracy, generalizability, and reproducibility. Addressing these data infrastructure challenges is essential to harnessing generative AI for accelerating discovery, characterization, and optimization of novel bioactive compounds from natural sources.
Introduction to Specialized Metabolites in Nature
Bacteria, fungi, plants, and animals produce a variety of specialized metabolites, including peptides, polyketides, sugars, terpenes, and alkaloids. These natural products play crucial roles in complex interorganism interactions, acting as signals, weapons, nutrient scavengers, and stress protectors. Although previously commonly used as antibiotics, chemotherapeutics, immunosuppressants, and crop protection agents, natural products have become less popular in industry in recent years than before due to the rise of combinatorial chemistry and high-throughput screening.
Biosynthetic Gene Clusters: A Pathway to Drug Discovery
The genes for most metabolite biosynthetic pathways in bacteria and fungi (and some plants and animals) occur as clusters in the genomes of the producing organisms: more than 2,500 of these biosynthetic gene clusters (BGCs) and their products have now been experimentally identified Characterized. This physical clustering has the potential to facilitate the identification of millions of putative new molecular biosynthetic pathways through computational genome analysis, providing a starting point for drug discovery.
The Role of AI in Predicting Biosynthetic Pathways
AI is currently being used to predict the chemical structure of BGC products based on DNA sequences, and key training data can be obtained through known biosynthetic pathways and their natural products. However, there is an urgent need for more efficient methods to filter and prioritize the large predicted biosynthetic diversity of natural products to identify drug leads.
Figure 1: Applications of artificial intelligence in natural product and drug discovery
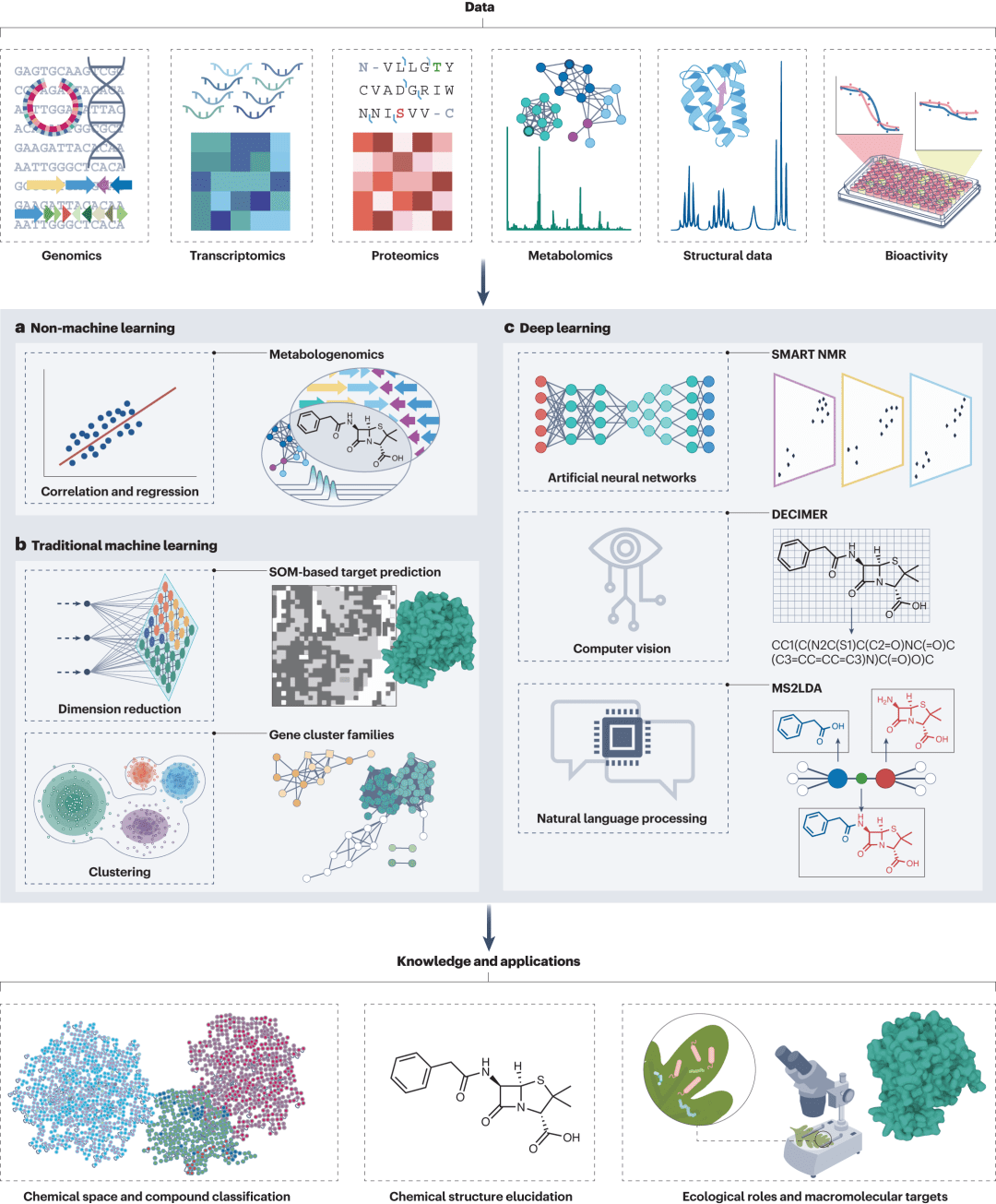
Figure 2: Example of natural product molecules discovered using AI
Including using the chemprop algorithm to discover the new antibiotic Halicin; using a convolutional neural network to predict the structures of rivulariapeptolides and symplocolide A from complex microbial extracts; using SVM to discover Prstinin A3 by mining whole-genome information.
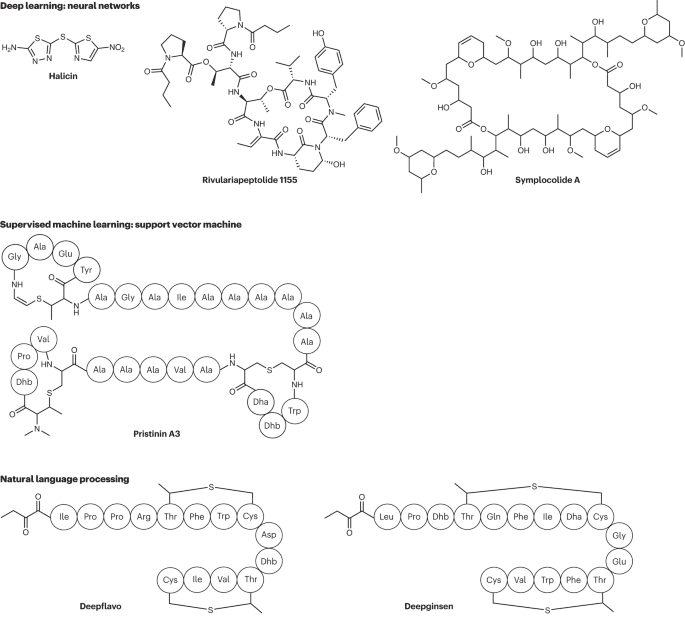
Figure 3: Prediction of bioactive and macromolecular targets based on genomic, metabolomic, and phenotypic data
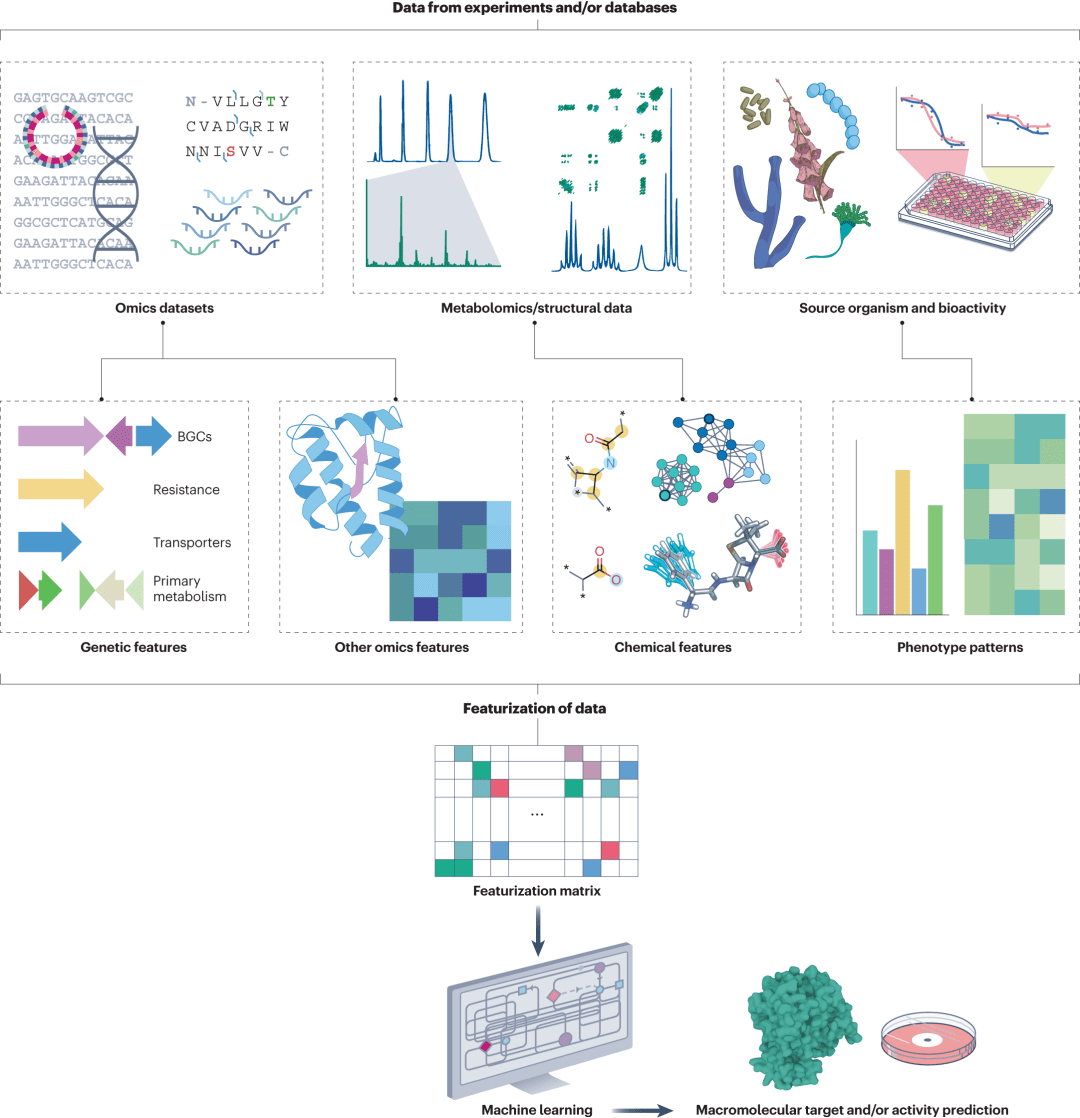
Figure 4: Molecular characterization of commonly used natural products, including pharmacophore, molecular fingerprint, SMILES, 3D dynamics and intermolecular interactions
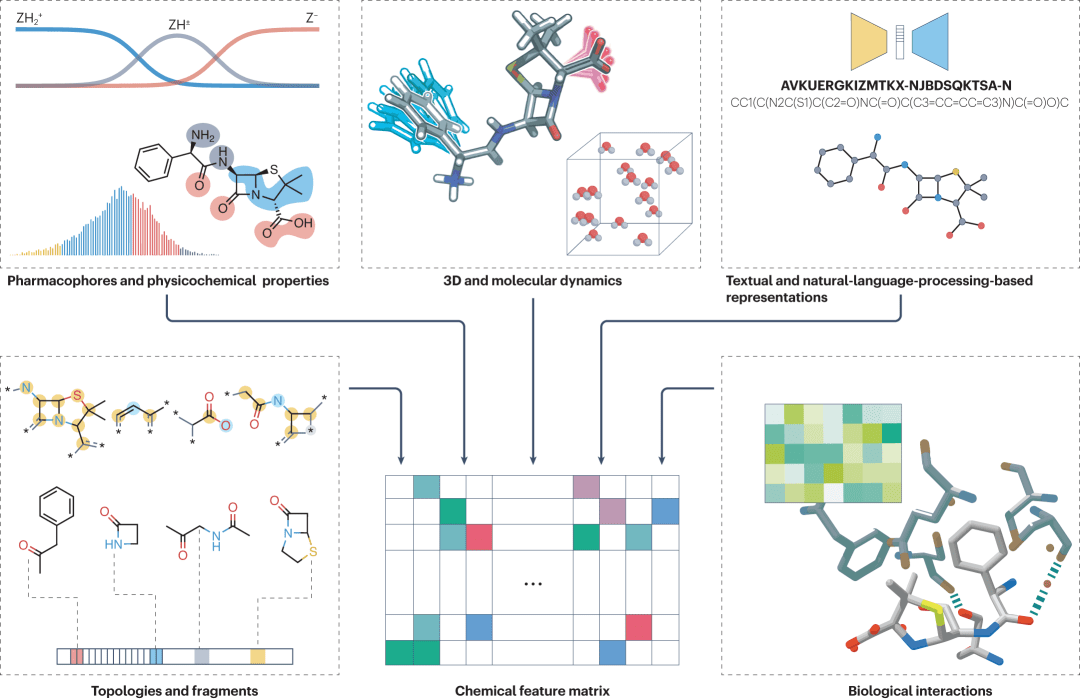
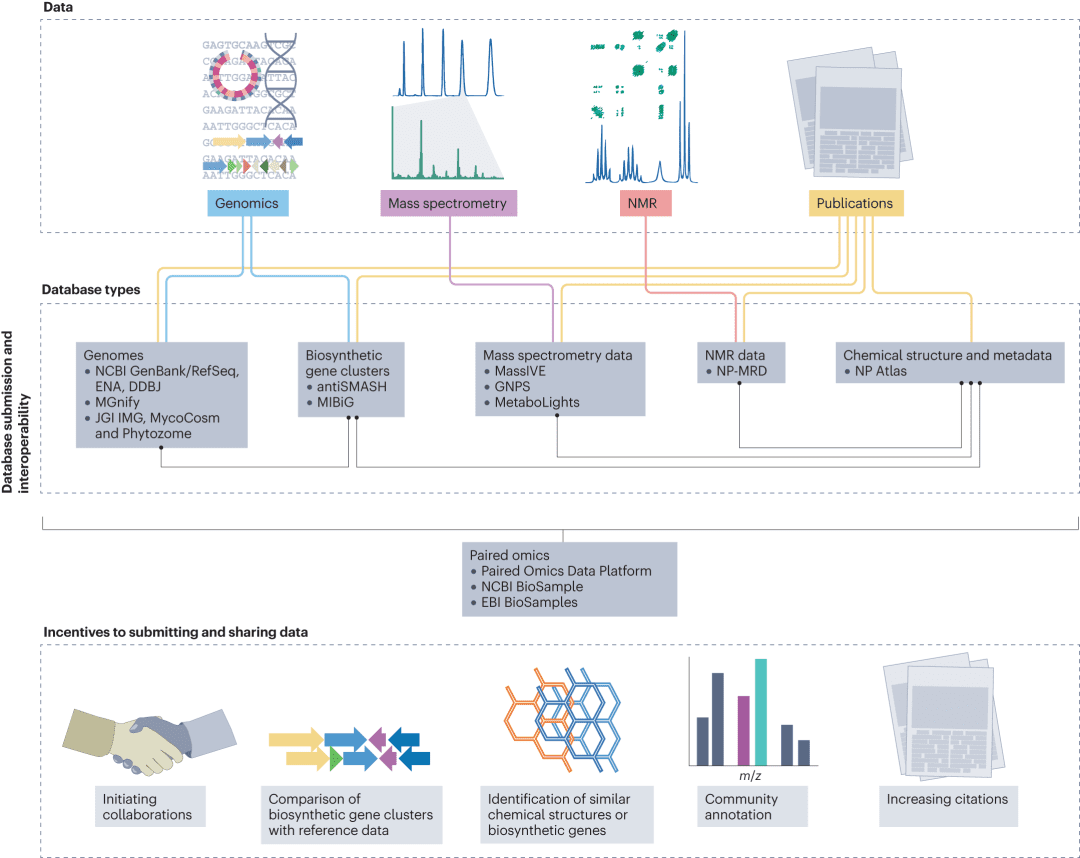
Figure 5: Storing and sharing natural product data: infrastructure and incentives